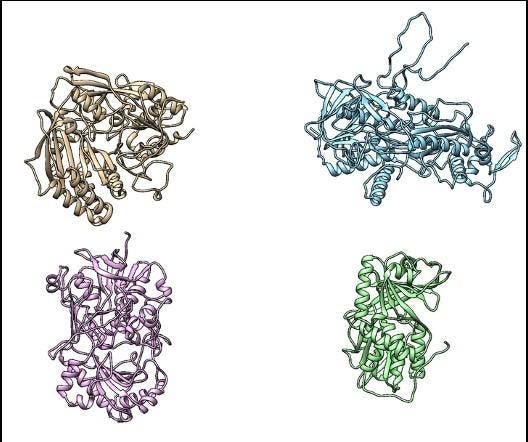
The three dimensional shape of proteins is determined by it’s sequence of amino acids
Image provided by bitBiome
Enzymes are proteins that guide (catalyze) very specific chemical reactions. They play indispensable roles in all life forms. Some enzymes carry out the day-to-day business of biology like digesting food, some are key for growth, some are for protection against toxins, and some of them give an organism a unique “super-power.” For instance, there is an enzyme in plants called RuBisCO which allows them to turn the sun’s energy into the basic building blocks for food, namely sugar. There are enzymes in certain bacteria that allow them to convert nitrogen gas from the atmosphere into the usable forms needed to support all life on earth. While humans make different enzymes of their own, for thousands of years we have found ways to utilize enzymes from microorganisms, long before they were even identified. Originally this was done by combining flour or grapes with naturally occurring yeasts or bacteria found growing in nature, to produce fermented products like wine or yogurt. However, in the last century we have learned how to isolate and produce the enzymes themselves and use them directly in these processes. For instance, “amylase” enzymes, typically found in saliva and used to start breaking down the starches we eat, are used to modify the starch in baking flour so that it does not rapidly crystallize and make the bread “stale.”
An enzyme called amylase has enabled sliced bread to be enjoyed much longer after purchase without becoming stale
getty
Natural microbes (bacteria and fungi) are a rich source of useful enzymes, but until recently only a tiny fraction of that potential diversity was accessible because it was necessary to be able to grow the organism in the lab to study it and understand what capabilities it might have. The world is full of organisms that can’t be grown in the lab, at least not without very specialized techniques. That is particularly true for organisms that live in very unusual settings like deep sea heat vents or in extremely salty water or soils. The general term for the organisms that live in these harsh environments is “extremophiles” and they are of interest because their enzymes have evolved to be able to “operate” under unusual conditions such as high temperatures or very acidic environments.
Novel Discovery Technology
There is a company based in Japan called bitBiome that has developed a new way to discover “super power” enzymes that are made by extremophiles or any other exotic category of microbes, without ever needing to grow that organism in a laboratory setting. Their technology begins with a microscopic sorting technique that allows them to isolate single bacterial cells from a mixed environmental or biological sample. They can then use recently developed technologies to make many copies of that bacterium’s DNA and sequence it, all without having to grow more of the isolated bacteria. Then they use a combination of evolutionary genetics, bio and chem-informatics and artificial intelligence to compare the sequences and predict three dimensional characteristics of these new proteins to similar information that is available in public and private databases that include known sequences or proteins of interest. The sequences that bitBiome collects are retained by their database, which now houses about 2.4 billion sequences from over 1 million organisms. With their sequencing technology, this database grows by over 1 billion sequences a year. AI is instrumental in helping to identify seemingly different enzymes with similar target activities.
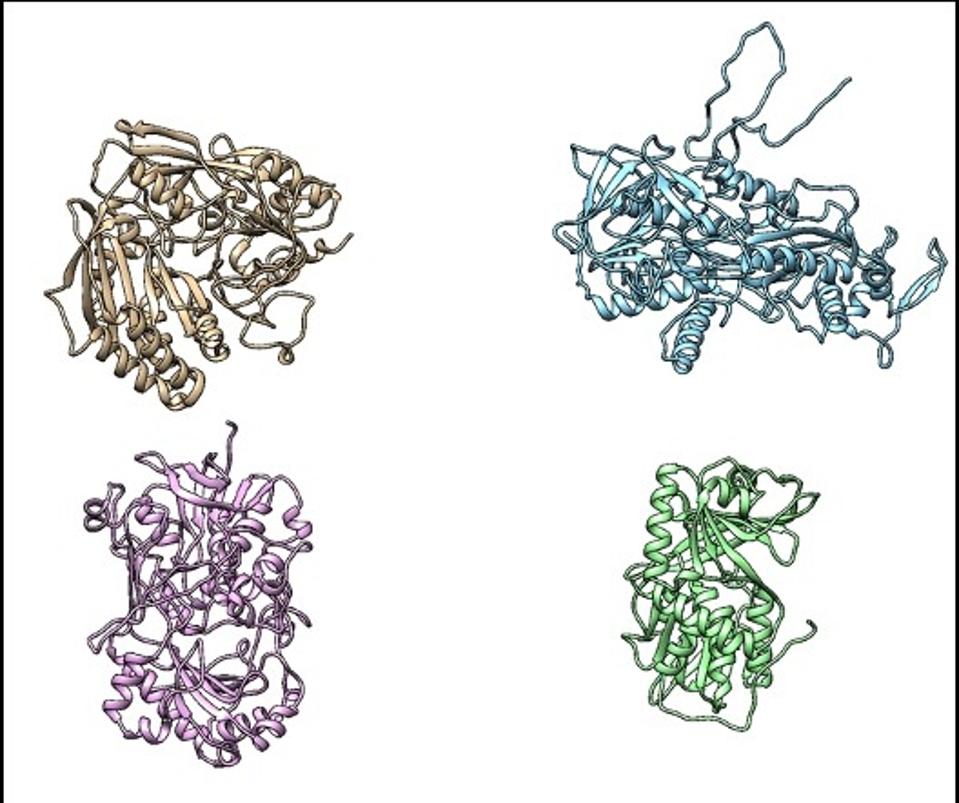
Proteins have intricate three dimensional structures that are determined by the sequence of amino acids that are coded by the DNA in a gene
Image provided by bitBiome
This is because enzymes from different organisms will sometimes look different, either in amino acid sequence or in 3D structure, but be able to carry out similar reactions. This allows bitBiome researchers to identify novel genes to insert into a standard laboratory protein-production organism and generate samples of potentially interesting new versions of the enzymes or proteins encoded by the isolated gene sequences.
The fit for a specific organic chemical compound as predicted by A-I based on the amino acid sequence of a protein
Image provided by bitBiome
This also allows them to evaluate the function of these newly identified enzymes or proteins. Even with AI, this discovery and “mining” process would be impossible without the ability to sequence individual cells. This is because common techniques rely on a decades-old sequencing technology called shotgun metagenomics, that frequently misses bacterial populations from complex samples. As a result of the shortcomings of this incumbent technology, we still have yet to identify over 99% of the bacterial species found on our planet.
Applications of the technology
There are many potential applications for this new enzyme discovery method including biomanufacturing, waste or toxin remediation, sustainable recycling of textiles and plastics, and the production of natural food additives, While bitBiome is currently focused on partnering with companies looking for new or improved enzymes to develop, they also have an internal product pipeline where they are developing their own suite of enzymes, proteins, or ingredients for commercial applications. bitBiome also offers their single cell sequencing to companies and academic researchers looking to analyze microbial populations at the single-cell level. This kind of research is critical, particularly in the areas of understanding the gut microbiome, but also for the understanding of complex microbial populations in soil, for example, among roots of growing crops. Bacteria have long been understood to interact with their environment in both negative and positive ways, from H. pylori, the gut bacteria implicated in gastric ulcers, to Lactobacillus, known to aid gut health.
The bitBiome technology to allow sequencing of single cells is a valuable new tool for the study of the human gut microbiome
getty
Partnerships
bitBiome has already found success with a number of partners including Ajinomoto, a Japanese company that has been providing enzymes to the food processing industry for many decades. Ajinomoto’s food enzyme technologies are contributing to better food productivity, cost reduction, and more efficient use of food resources. The collaboration between bitBiome and Ajinomoto is focused on using bitBiome’s proprietary technology to discover entirely new enzymes with novel functionalities and applications where there have not been adequate solutions to date.
It will be fascinating to see what new functions and capabilities can be discovered and how they can be employed by the growing bioeconomy.